DNA Repair - Definition, Types
DNA REPAIR
Definition:
Restoring the damaged part of DNA is called DNA repair. DNA
is a fragile molecule that can be easily damaged. It is prone to damage due to
replication and mutagenic agents. The following damages are caused to DNA.
I. Insertion of a wrong base during replication. Eg. Conversion
of cytosine to uracil (found only in RNA). UV rays alter bases.
2. Loss of a purine base.
3. Alteration of bases. Eg. amino group (deamination)
converts C to U.
4. Breakage of DNA strands. It may be single stranded break
(SSB) or double stranded break (DSB).
5. Covalent linkages between bases. It may be on the same strand
(intrastrand) or between strands (interstrand).
6. Mismatches of the normal bases. Eg. Incorporation of U
(found only in RNA) instead of T.
The following agents cause damage to DNA:
1. Radiation - UV rays, X rays, gamma rays, etc.
2. Highly reactive oxygen - radicals produced during normal
cellular respiration.
3. Chemicals in the environment (Hydrocarbons) including those
found in cigarette smoke.
4. Plant and microbial products Aflatoxins.
5. Chemicals used in chemotherapy especially chemotherapy of
cancer.
If the DNA damage is not repaired, it will lead to mutation.
Thus DNA damage may cause mutation; if not corrected. A failure to repair DNA
produces mutation. The following are the DNA repair mechanisms: s seen in
living cells
1. Direct repair
2. Excision repair
3. Mismatch repair
4. Recombinational repair.
1. Direct Repair
In direct repair mechanism, the changes introduced in the
bases within a DNA are corrected directly without breaking the DNA. Here the
repair is done without breaking the backbone of DNA. It is carried out by only
one enzyme. Ultraviolet rays cause thymine dimers resulting in the linking of
intrastrand thymines. It is corrected by direct repair. The thymine dimers are
corrected by the process of photoreactivation. Photoreactivation is the
reversal of DNA damages caused by UV rays by the visible light. The repair is
carried out by the enzyme DNA photolyase.
This enzyme binds to thymine dimers in dark. When the cells are exposed to sunlight, folic acid absorbs the light and uses the energy to break the intrastrand linkages of thymine dimer.
2. Excision Repair
The damaged base or bases of DNA are excised and the gap is
filled with the correct base or bases. Excision repair is a dark repair
enzymes. and is independent of light. It requires a set of three or four. The
excision repair is of two types. They are the following:
1. Nucleotide
excision repair
2. Nucleotide stretch excision repair
1. Nucleotide excision Repair:
In nucleotide excision repair, single damaged base is
removed with the backbone of DNA and is replaced by a correct one. It involves
the following steps:
1. The enzyme glycosylase recognizes the damaged base
(deami- nation) and binds to it on the DNA strand.
2. It hydrolyzes the N-glycosyl bond linking the base to the
sugar-phosphate backbone. The damaged base is excised and re- moved. This
creates a baseless site in the DNA strand.
3. Then an endonuclease and phosphodiesterase hydrolyze the phosphodiester
bond so that the sugar and phosphate are removed.
4. Then DNA polymerase fills up the missing nucleotide.
5. The DNA ligase seals the nick.
2. Nucleotide Stretch Excision Repair:
In nucleotide excision repair, a damaged segment of DNA is
excised and replaced by the correct segment. It involves the following steps.
1. The enzyme endonuclease is attached to the damaged site.
Then the enzyme cuts the polynucleotide on either sides of the dam- aged site
resulting in the excision of a segment of 12 nucleotides including the damaged
part.
2. The enzyme 5'-3' exonuclease removes the excised segment
by breaking the bond attaching it to the complementary strand.
3. The gap is filled by DNA polymerase I.
4. The nick is closed by a phosphodiester bond formed by DNA
ligase.
3. Mismatch Repair:
Mismatch is a wrong incorporation of nucleotide in the newly
synthesized strand. It is an error in replication. Mismatch repair is an
excision repair independent of light. It is a dark process. It is a post
replicative repair.
During replication, the nucleotides of DNA are copied in the
daughter strand in 100% accuracy. However very rarely an incorrect nucleotide is
incorporated as a result of a shift in the electronic state of the nitrogen
base of an incoming nucleotide or of the template nucleotide. As a result, A
may pair with C instead of Tor G may pair with T instead of C. As A,T,G,C are
normal components of DNA, mismatch repair systems need some way to determine
whether, a particular base is the correct base at the given site. The repair
system identifies this, by comparing the template strand which contains the
original nucleotide sequence and the strand containing the wrongly incorporated
base. This distinction can be made based on the pattern of methylation in newly
replicated DNA. In E.coli, the A in GATC sequence is methylated subsequent to
its synthesis.
Following replication, one strand remains methylated and the
other remains unmethylated till methylase acts on this new strand to bring
about methylation. Thus a time interval occurs during which the template strand
is methylated and the newly synthesized strand is unmethylated.
The mismatch repair system uses this difference in
methylation state to excise the mismatched nucleotide in the nascent strand and
to replace it with the correct nucleotide by using the methylated parent strand
of DNA as template. proteins, namely In E.coli, the mismatch repair system
needs four mutH, mutL, mutS and mut U.
The mutS protein recognizes the mismatches and binds to them to initiate the repair process; mutH and mutL proteins join with muts. The mutH has endonuclease activity and it excises the mismatch segment with a 1000 or more nucleotide length. Exonuclease removes the excised segment. DNA polymerase III synthesizes a new segment and fills the gap. DNA ligase seals the nick.
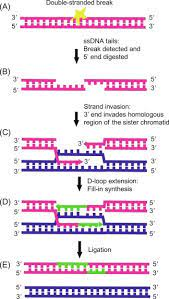
4. Recombinational Repair:
In recombinational repair, the damaged part of a DNA is replaced by a part of another DNA molecule. This repair mechanism is found in E.coli and virus. Recombinational repair requires the presence of an identical sequence similar to the damaged sequences. The enzymatic machinery responsible for this repair process is nearly identical to the machinery responsible for chromosomal cross over during meiosis. It is a post-replicative repair mechanism. DNA dimers, caused by UV rays, are repaired by photoreactivation or excision repair mechanism. When DNA polymerase-III reaches the dimer (thymine dimer), replication is blocked. Polymerase III starts reacting at the dimer site and starts its proof reading role. If the dimer is not repaired by photoreactivation or excision repair mechanism, the DNA polymerase- III cannot make complementary strand for five seconds. However, DNA polymerase-III restarts DNA synthesis, in post dimer initiation site by skipping over the dimer segment (lesion). This leaves a large gap of 800 bases in the newly synthesized DNA strand. An error-free segment of the DNA of the fork is excised and inserted in place of the gap created by thymine dimer. Rec B cuts out an error free segment from the similar DNA and rec A exchanges the segment to the gap in the daughter strand. The gap if any in the DNA is filled by DNA polymerase I and joined by DNA ligase. The gap in the donor strand is likewise filled up by the same enzymes.





Comments